CDR Tickets
| Issue Number | 4523 |
|---|---|
| Summary | [Term] Remove Legacy-combination chemotherapy terms from the CDR |
| Created | 2018-09-19 12:22:43 |
| Issue Type | Inquiry |
| Submitted By | Osei-Poku, William (NIH/NCI) [C] |
| Assigned To | Kline, Bob (NIH/NCI) [C] |
| Status | Closed |
| Resolved | 2019-02-20 09:37:26 |
| Resolution | Fixed |
| Path | /home/bkline/backups/jira/ocecdr/issue.233310 |
Please remove all terms that have a TermType of “Legacy-combination
chemotherapy”
and a SemanticType of “drug/agent combination” from the CDR. These are
terms that were created specifically for indexing some clinical trials
in the past. we stopped using them several years ago. They create a lot
of noise in search results and on reports.
Step 1: make sure nothing links to these terms.
Step 2: submit remove job to GateKeeper for the term
docs
Step 3: "delete" the docs
For step 1:
SELECT DISTINCT d.id, d.title, q.path
FROM query_term q
JOIN document d
ON d.id = q.doc_id
WHERE (q.path LIKE '%cdr:ref'
OR q.path LIKE '%cdr:href')
AND q.int_val IN (
SELECT t.doc_id
FROM query_term t
JOIN query_term s
ON s.doc_id = t.doc_id
WHERE t.path = '/Term/TermType/TermTypeName'
AND t.value = 'Legacy-combination chemotherapy'
AND s.path = '/Term/SemanticType/@cdr:ref'
AND s.int_val = 256171) -- Drug/agent combinationresults in no hits on DEV. There's lots of hits on PROD, but they're all for protocol documents (confirmed by adding this to the query):
AND q.path NOT LIKE '/InScopeProtocol%'
AND q.path NOT LIKE '/CTGovProtocol%'For steps #2 and #3, we can use the following query, which picks up 2,718 documents on both DEV and PROD. I will create the hotfix/remove job to remove these docs from GateKeeper on DEV.
SELECT DISTINCT t.doc_id
FROM query_term t
JOIN query_term s
ON s.doc_id = t.doc_id
WHERE t.path = '/Term/TermType/TermTypeName'
AND t.value = 'Legacy-combination chemotherapy'
AND s.path = '/Term/SemanticType/@cdr:ref'
AND s.int_val = 256171 -- Drug/agent combinationRemove requests have been sent to gatekeeper-blue-dev
for all 2,718 documents, and their active_status value has
been set to D in the CDR. Any idea why the push job doesn't
seem to be making any progress, ~volker? (See http://gatekeeper-blue-dev.cancer.gov/admin/RequestHistory/RequestHistory.aspx.)
Yep! Because the remove job is #2 in the processing queue. #1 is the job that is processing the 15k media documents and it's running pretty slowly. There are about 12k documents to go. It'll certainly run all through the night.
The documents have been deleted on QA. The hotfix-remove job on QA didn't do anything, as none of the documents were in pub_proc_cg on that tier.
The terms still come up in searches on QA and DEV.
I looked at CDR373802, and I don't see "Legacy-combination chemotherapy" anywhere.

Please also remove all terms with a semantic type of “drug/agent combination”. We failed to make this clear in the requirements.
I increased the priority as we found out that some of these terms are affecting the vendor output and we would want to get them fixed as soon as possible.
What about terms that have a TermType of “Legacy-combination chemotherapy” but do not have a SemanticType of “drug/agent combination”? I came away from our last discussions of this ticket that those were supposed to be deleted as well. Am I mis-remembering? I thought you had been saying the new requirements were supposed to call for deleting:
documents which meet only the first condition
documents which meet only the second condition
documents which meet both conditions
(in contrast with the original requirements, which only called for deleting documents which meet both conditions).
Documents that have a TermType of “Legacy-combination chemotherapy” without a SemanticType of “drug/agent combination” should be removed as well. I had the initial impression that all of the terms would have a SemanticType of “drug/agent combination”. Essentially, we do not need any terms that have a SemanticType of “drug/agent combination”. Neither do we need any term that has a TermType of “Legacy-combination chemotherapy”. Hope this clears up the confusion. Sorry about that.
I'm going to use the following query on DEV to implement the new requirements:
SELECT DISTINCT doc_id FROM (
SELECT doc_id
FROM query_term
WHERE path = '/Term/TermType/TermTypeName'
AND value = 'Legacy-combination chemotherapy'
UNION
SELECT doc_id
FROM query_term
WHERE path = '/Term/SemanticType/@cdr:ref'
AND int_val = 256171 -- Drug/agent combination
) AS double_queryIt picks up 782 documents on DEV (because we're already deleted the documents which met the original requirements). The query matches exactly 3,500 documents on PROD.
All the documents which match the new requirements have been removed from GateKeeper/Cancer.gov on DEV-RED (we're hooked up to a different color on DEV than we were when we tested using the original criteria) and deleted in the CDR on DEV. Please verify.
As a side note, gatekeeper-dev-red was much more cooperative than gatekeeper-dev-blue.
Mary has reviewed all the terms retrieved by the query and confirmed that they can all be deleted.
~bkline When you have a little time to spare, could you please apply this fix on QA ? 🙂 We changed the priority to critical about 2 weeks ago.
Done on QA.
Verified on QA. Ready for PROD. Thank you!
782 documents removed on QA
3500 documents being removed on STAGE
PROD will be done as soon as the STAGE removal is complete
~volker and ~learnb: I'm running into a snag on STAGE. I can see from http://gatekeeper-stage.cancer.gov/admin/RequestHistory/RequestHistory.aspx that the hotfix-remove push job has finished on GateKeeper. However, although the scheduler has been asking GateKeeper for the confirmation of that completion every 5 minutes (plus one manual kickoff of the verification task), GK still isn't giving us a completed status. Is GateKeeper in a bad place on STAGE?
No, STAGE is fine and the job finished successfully.
I would guess that the scheduler is the one acting up again.
I'm wondering now if the problem may be that you ran a Hot-fix instead of a Hotfix-remove. We so rarely run remove jobs that I don't know on top of my head what the difference is.
Well, the logs record that I'm asking GK for the verification every 5 minutes. See d:/cdr/logs/push-job-verifier.log. So it would appear that the scheduler is healthy enough to run the jobs.
No, I ran a hotfix-remove.
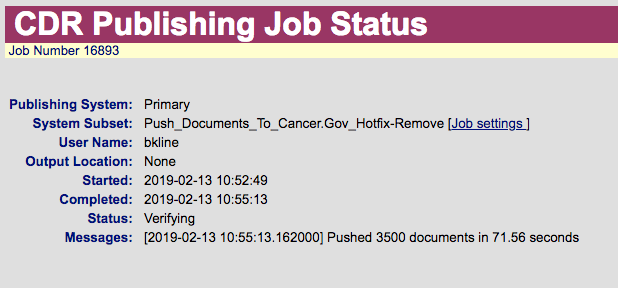
OK, I only saw the 'Hotfix' listed on the GK side.
Documents removed from cancer.gov and marked as deleted in the CDR on STAGE and PROD.
Actually, the CDR document deletion jobs are still running (on both tiers).
Now I remember why we were holding off on this ticket. We still haven't removed the protocol documents on STAGE and PROD, and many of these terms have links from those protocols. Apparently we have never used the admin tool for marking documents as deleted for documents which have incoming links: there is at least one bug in that tool which fails when there are inbound links still present, and that path has never been hit before now.
We need to get Lakshmi to weigh in on the decision as to whether to proceed with the streamlining task. It would be ideal if we could take care of getting that approval before we begin work on the next release so we don't have all this stuff still hanging out there.
The current status of this ticket is that all 3,500 terms have been removed from GateKeeper on STAGE and PROD, but only 1,107 of have been deleted (on both tiers) because of the incoming links.
I have attached a log from my analysis of the links to the documents which are yet to be deleted. I did the analysis on STAGE, but I expect PROD to be the same.
I figured out a way to delete the documents from my DEV VM with a custom script. I have run the script against STAGE. If after reviewing that tier (and the list of incoming links I attached) you conclude that STAGE looks good I'll run the script against PROD. (I had to run another hotfix-remove publishing job, because the Term documents were still active, so they got published again.)
Looks good on STAGE. Please proceed to run the script on PROD. Thank you!
Done on PROD. Please verify.
Verified on PROD. Thank you!
| File Name | Posted | User |
|---|---|---|
| Legacy-Combination Advanced search.JPG | 2018-11-14 16:01:18 | Osei-Poku, William (NIH/NCI) [C] |
| Legacy-Combination search.JPG | 2018-11-14 16:01:17 | Osei-Poku, William (NIH/NCI) [C] |
| ocecdr-4523.out | 2019-02-13 16:23:27 | Kline, Bob (NIH/NCI) [C] |
| screenshot-1.png | 2019-02-13 11:30:08 | Kline, Bob (NIH/NCI) [C] |
| Screenshot 2018-11-15 06.51.10.png | 2018-11-15 06:51:31 | Kline, Bob (NIH/NCI) [C] |
Elapsed: 0:00:00.001217