CDR Tickets
| Issue Number | 4598 |
|---|---|
| Summary | [Summary] Links to Clinical Trials Report |
| Created | 2019-03-26 15:15:18 |
| Issue Type | New Feature |
| Submitted By | Osei-Poku, William (NIH/NCI) [C] |
| Assigned To | Englisch, Volker (NIH/NCI) [C] |
| Status | Closed |
| Resolved | 2019-08-12 12:25:55 |
| Resolution | Fixed |
| Path | /home/bkline/backups/jira/ocecdr/issue.241966 |
We talked a while ago about getting a report that would let us know the number of links in summaries (Protocol Refs) that are linking to trials in Cancer.gov vs trials in Clinicaltrials. gov. We probably want to start with getting an ad hoc query listing the summaries with protocol ref links and whether they link to Cancer.gov trials or Clinicaltrials.gov trials. We also want to know the count of the links as well. If there is a future need for a real CDR report, I would put in a ticket with specs for that.
We probably want to start with getting an ad hoc query listing the summaries with protocol ref links and whether they link to Cancer.gov trials or Clinicaltrials.gov trials.
Yes, we can create an ad-hoc query listing the summaries with protocol ref links but it's impossible to identify whether these refs link to Cancer.gov or Clinicaltrials.gov because that information is not stored anywhere in our database. As you remember we modified all ExternalRefs pointing to clinical trials and the related filter so that all ProtocolRef elements are now pointing to
https://www.cancer.gov/clinicaltrials/<NCT-ID>It is Cancer.gov who identifies where the protocol lives and redirects the link accordingly.
We also want to know the count of the links as well.{quote}
Is that the count per summary or the total count?
let us know the number of links in summaries (Protocol Refs) that are linking to trials in Cancer.gov vs trials in Clinicaltrials. gov.{quote}
There are actually three options available:
links pointing to Cancer.gov
links pointing to ClinicalTrials.gov and
links pointing nowhere. These are links that were removed by our filters due to blocked documents but are listed as ProtocoRefs
How would you like to have the third option handled?
These links may need to be cleaned up from the summaries. Please display that information in a separate column.
I'm attaching a screenshot of the report I've created so far. I'm not certain what extra information you're gaining by creating an extra column for the ProtocolRefs that aren't linking anywhere on Cancer.gov. My question was more geared towards "What would you like to display if not None"?
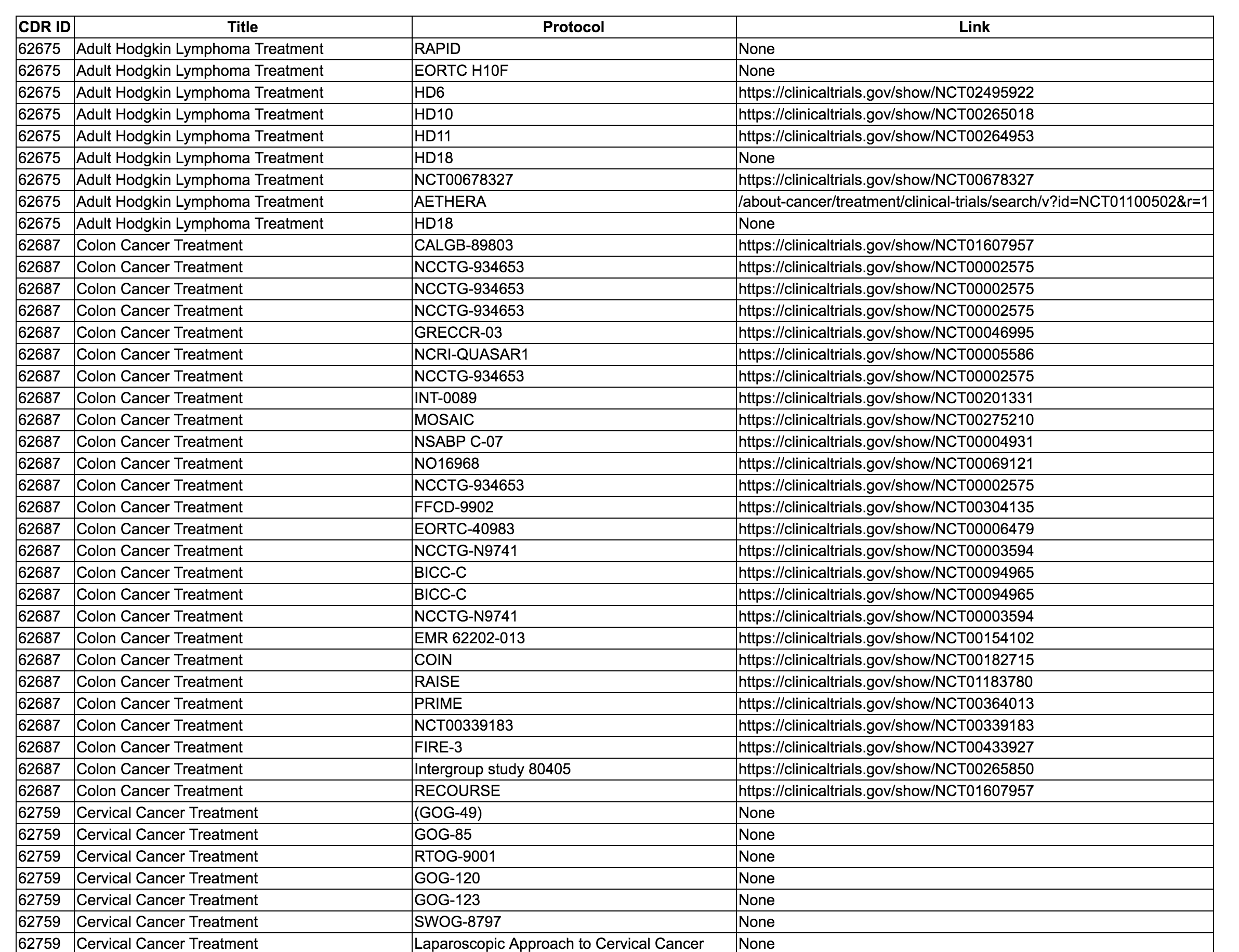
I think None is fine. Thanks!
For myself to remember:
I will need to re-index the summaries after adding the attribute nct_id to the query_term table.
I've updated/created the following files:
SummaryAndMiscReports.py
SummaryProtocolRefLinks.py
For the moment I'm limiting the report to only display results for 10 summaries. The full report takes several minutes to complete.
I created the new Summary Report option Summaries with ProtocolRef Links
Please have a look and let me know what you think.
I am getting a "Session not active" error message when I click on the link.
Please go through the Admin Reports interface instead of using the link from the above comment. I copy/pasted the name from the menu page but didn't mean for JIRA to make this a link.
Sure. What is the name of the report on the menu? I didn't know it was on the menu.
It's "Summaries with ProtocolRef Links"
Found it thanks!
The report looks good to me. I see that for the cancer.gov links you are including only the relative links/urls instead of the FQDN. Is it because of space ?
No, that's the output that the tool I'm using returns. I'm guessing that the tool drops the hostname for local URLs. I can add that to the report if you'd prefer.
Yes, please! Add it. Thanks!
I've modified the report to display the full URL and I removed the restriction for only 10 documents.
Please note that this report will run for a long time (around 7 minutes). I've added a note to that affect to the menu option. Obviously, PROD is a bit faster than DEV but I'm guessing the report will still run around 4-5 minutes.
I cannot see the report on the admin menu anymore.
Please remember that Bob had updated DEV with the Joule release last week. This change is not merged into Joule yet. I'll have to manually restore the files and once you approved the report I'll move it into Joule.
Give me a few minutes to make the change before testing again.
~oseipokuw, the report is available on DEV again.
This looks good on DEV. Would you be able to provide stats for the report? That is a count for each of the categories (Cancer.gov, Clinicaltrials.gov, and None).
Do you want those stats as part of the report or just a general count right now?
If you want the counts listed as part of the report where would you like them displayed (bottom/top)?
Please include it as part of the report and at the top left corner of the report. Thanks!
I've not been able to add the table with the counts to the left but it displays at the top. I will have to check with Bob if it's possible to position the tables in the HTML format.
SummaryProtocolRefLinks.py
[cdr4598-prot-links 3b047f0f]
This change is ready for review on DEV.
Looks good. Thanks! There is no need to align the stats table to the left of the page.
Verified on DEV.
The changes have been merged into our release branch Joule.
The report times out on QA (and now on Dev even though it worked on DEV before).
Unfortunately, the QA server is our slowest machine and the timeout is right around the report completion time. The STAGE and PROD servers are somewhat faster but it's still possible we're running into the timeout on occasion. I'll check if there are ways to make the report faster. Otherwise, we'll have to rewrite the report to be run as a batch job.
I tried several different approaches to speed up the report. The first approach did not improve much. My second attempt is removing duplicates from the list of ProtocolRefs so that we're only testing the URL once for each summary. This seems to decrease processing time from about 15 minutes to about 11 minutes - about 25% improvement. If this isn't working we'll have to rewrite the report to make it a batch report.
Please go ahead and test this version on DEV. If this report will work better I'll have to check with Bob if it's possible to modify the build for Joule or if we need to push the ticket to Kepler instead.
The URL for the modified report is
https://cdr-dev.cancer.gov/cgi-bin/cdr/SPR2.py?Session=guest
The report did run successfully this time around. Thanks!
I've copied the report to QA for testing:
https://cdr-qa.cancer.gov/cgi-bin/cdr/SPR2.py?Session=guest
If you're able to run it here as well, I will replace the original report with this version.
The report is failing on QA (and DEV, even though it worked on DEV a couple of days ago).
You were running the report SPR2.py and not the original SummaryProtocolRefLinks.py, right?
It is too bad that the report still times out but the report still depends on how fast a requests to CT.gov and Cancer.gov get back. The new report has only about 1,500 links to test while the old version of the report was testing about 2,200. I was hoping this change was good enough to keep the report execution within the 15 minute limit.
I'd suggest to submit a new ticket for Kepler to convert the report into a batch report. Since the report execution time is largely dependent on the network speed I don't think it would complete on the faster PROD server. The report will still finish on occasion - it hasn't failed for me so far - but it's not reliable.
Yes, I used the links you provided above. I just tried it again and it timed out again.
I'd suggest to submit a new ticket for Kepler to convert the report into a batch report. Since the report execution time is largely dependent on the network speed I don't think it would complete on the faster PROD server. The report will still finish on occasion - it hasn't failed for me so far - but it's not reliable.
Will do. Thanks!
Based on William's experience that the report is still timing out for him, we decided to revert my first approach - using the Python module grequests in favor of the module requests - and using the original Python module. I am still removing duplicate links within each summary.
The changes have been tested on DEV and the updated report is creating the same results. Changes have been copied to the Joule release.
Verified on QA!
Verified on PROD. Thanks!
| File Name | Posted | User |
|---|---|---|
| Screen Shot 2019-06-13 at 10.52.08.png | 2019-06-13 10:55:12 | Englisch, Volker (NIH/NCI) [C] |
Elapsed: 0:00:00.001291