CDR Tickets
| Issue Number | 5093 |
|---|---|
| Summary | Importing multiple terms from the NCI Thesaurus |
| Created | 2021-08-11 18:10:48 |
| Issue Type | New Feature |
| Submitted By | Osei-Poku, William (NIH/NCI) [C] |
| Assigned To | Kline, Bob (NIH/NCI) [C] |
| Status | Closed |
| Resolved | 2022-01-21 16:11:23 |
| Resolution | Fixed |
| Path | /home/bkline/backups/jira/ocecdr/issue.296357 |
I am creating this ticket as a placeholder for possible discussions on Thursday about importing multiple terms from the NCI Thesaurus into the CDR for review and publishing to the Drug Dictionary. Requirements will come later after we've had the chance to determine what is possible with the new EVS RESTAPI.
Hi ~bkline I am wondering if you could provide us with sample copies of some of the files we import from EVS. Any of them should be good or you can provide for these:
CDR0000043234 - C2039
CDR0000355725 - C91702
CDR0000792379 - C147030
CDR0000797414 - C158750
We don't actually import files from the EVS, we extract specific pieces of information from responses we get to API requests and import those values into the CDR. I have captured such a response, corresponding to the concept IDs you identified, and attached it to the ticket. Will that meet your needs?
Yes, this is helpful. Thank you!
Mary and I have looked at the data from the responses but we are not able to identify any specific criteria for importing the terms we need for the drug dictionary. Essentially we need to import
1. Drug terms
2. That are cancer related that we don't already have and download them for review and import into the CDR.
3. If we already have the terms, then to check to see if there are any updates for review and import them.
In nearly all cases, the new cancer terms are CTRP provided terms so if we are able to concentrate on only terms provided CTRP, we will get all the terms we need for the drug dictionary. Just for your information, Lori, provided us with an FTP site (https://evs.nci.nih.gov/ftp1/CTRP/) that has all the CTRP terms and it appears to be up to date but the definitions are not stored there.
I spoke with Mary about this and we agreed that using the
"source": "CTRP" and/or
_type": "Contributing_Source",
"value": "CTRP"_
elements should be good in identifying the terms she needs. However, it will help if it can further be limited to just drug terms, as it is possible that using these elements could still include diagnoses and other terms. I think eventually, having some test data to look at will be helpful.
Can you be more specific about what "test data" means in this context?
Since we have not been able to determine the selection criteria, I was thinking about when this is being implemented on DEV, we will be able to review the (test) data a bit more to ensure we are getting what we want.
I have retrieved all of the descendants of Pharmacologic Substance
(Code C1909) from the EVS, and all of the Term documents in
the CDR with a semantic type of Drug/agent, Drug/agent category, or
Drug/agent combination (though I didn't see any with this last semantic
type, which surprised me). I have attached an Excel workbook with four
sheets.
Drugs found in both systems, but with different names (many involving discrepancies in case)
Drugs found in the EVS, but not in the CDR
Drugs found in the CDR, but not in the EVS
Drugs found in the CDR, but without an NCI Thesaurus concept code
So the next steps, I think, include:
you review the data in the workbook
you tell me what other properties I need to compare, other than preferred name
you tell me how I can tell which drugs in the EVS "are cancer related" (see your September 9 comment above) — will fetching all 20,264 drug concepts from the EVS and looking for the CTRP source as described above be the same thing as filtering by "cancer-related"?
you tell me if fetching descendants of C1909 is the appropriate approach to finding drugs
We have reviewed the data and your questions as well but I wanted to mention here that from Lori's 9/22/21 email (I will attach to this ticket), it looks like we may be able to get the terms we need by retrieving terms with in the two subsets provided in the email and avoid coming up with a way to identify cancer-related terms. The subsets she mentioned in the email are:
1. NCI Drug Dictionary Terminology C176424
2. CTRP Agent Terminology C116978
Looking at the data you in the .json file above, it looks like the subsets can be found in the ("type": "Concept_In_Subset") element_._
There seems to be a third subset "relatedName": "CTRP Terminology" which she did not mention in her email so we may need to clarify with her.
"type": "Concept_In_Subset",
"relatedCode": "C116978",
"relatedName": "CTRP Agent Terminology"
Also, in the data, there is a second relatedName called "CTRP Terminlogy" which will also be helpful in retrieving but we may have to clarify with Lori because she didn't mention it in here email.
"type": "Concept_In_Subset",
"relatedCode": "C116977",
"relatedName": "CTRP Terminology"
The NCI Drug Dictionary Terminology C176424 subset can also be found in the .json data you provided.
"type": "Concept_In_Subset",
"relatedCode": "C176424",
"relatedName": "NCI Drug Dictionary Terminology"
},
In subsequent comments I will provide more information about our review and also attempt to answer the questions you had.
With regards to the spreadsheet, we are reviewing the No Codes tab to understand why there is such a large number of terms without the concept codes with the hope that once those terms are cleaned up, it may make it easier to also compare with the codes in addition to the preferred name.
With regards to the Name Changes tab, we wouldn't want to see changes only because of case differences because we follow a different case convention from EVS. What we would want to see is if the preferred name has changed, for example, the inclusion of a hyphen.
With regards the Missing from the CDR tab, we haven't looked
at every single one of the terms yet but there are some terms that we
found in the CDR.
|C205|Acyclovir|
39134|
|C212|Recombinant Corticotropin|
39133|
We haven't reviewed the Missing from EVS tab yet.
So the next steps, I think, include:you review the data in the workbook
Still reviewing.
you tell me what other properties I need to compare, other than preferred name
Can you please clarify this question? Are the results of the comparing found in the Name Changes tab or it includes the other tabs ?
you tell me how I can tell which drugs in the EVS "are cancer related" (see your September 9 comment above) — will fetching all 20,264 drug concepts from the EVS and looking for the CTRP source as described above be the same thing as filtering by "cancer-related"?
We hope that by using the subsets provided in the previous comments, there will not be the need to specifically find cancer related terms.
you tell me if fetching descendants of C1909 is the appropriate approach to finding drugs
Same as above.
Can you please clarify this question?
If I'm looking at a CDR Term document, pull out its
concept ID, find the concept record in the EVS with the same concept ID,
and determine that the concept names are different between the CDR and
the EVS I'll include that concept in a report of what's not in sync. Is
the concept/term name the only pair of values you want me to compare
between the two systems to determine what should be included on a report
of deltas? Similar question for an automated batch job to update
Term documents.
Thanks for the clarification!
We are looking for:
1. True difference in preferred name (not just capitalization or
punctuation)
2. Differences in definition beyond capitalizations and
punctuations.
3. Other Names that are present in one but absent in the other
(especially brand names)
With regards the Missing from the CDR tab, we haven't looked at every single one of the terms yet but there are some terms that we found in the CDR.
C205
Acyclovir
39134|
C212
Recombinant Corticotropin
39133|
Well, yes those documents are in the CDR. But they've got different concept IDs than the EVS has, so the software won't find them, will it?
Here's a report showing the difference between what the EVS has and what the CDR has for the drug terms they both have.
Thanks ~bkline This is a very useful report that we would like to keep it on the reports menu, if possible.
There are a few things we are thinking about doing with this information.
Please add the preferred names (from both EVS and CDR) to the report, perhaps highlighting them so they stand out from the rest of the data. We will do another review after you the preferred names to the report.
Can we run a global to update the CDR in cases where the EVS has other names that are not present in the CDR?
Can we run a global that will update the CDR in cases where there are true differences between the definitions?
I am working on a revised version of the report as requested, but I want to be sure you had noticed that the report you already have shows the preferred name whenever the CDR and the EVS don't match.

Also, in the data, there is a second relatedName called "CTRP Terminlogy" which will also be helpful in retrieving but we may have to clarify with Lori because she didn't mention it in here email.
"type": "Concept_In_Subset",
"relatedCode": "C116977",
"relatedName": "CTRP Terminology"
What was the outcome of those discussions with Lori about the possible inclusion of this term? If you do include that term for the "subset" retrieval from the EVS, the number of terms matched will balloon well beyond the number matched without it. With just the other two terms used for the subset matching, the number of concepts identified is in the same rough ballpark as the number of CDR Term documents found with semantic type of Drug/agent, Drug/agent category, or Drug/agent combination. Adding in "CTRP Terminology" results in a number of concepts which is several times the size of the set of CDR drug terms, and would render efforts to identify drugs which the EVS has but are missing from the CDR futile.
I've been holding off on modifications to the report pending the outcome of those discussions with Lori.
I have attached a report which shows CDR Term documents
on PROD which have a "drug/agent..." semantic type but which have no
NCIThesaurusConcept element. Of the 675 documents, 106 have
preferred names which match that of a concept in the EVS, as recorded in
the second column.
(By "match" I mean comparing with normalized whitespace and case.)
I should also point out that in some cases it appears that the wording in a definition has been improved or corrected in the CDR, and automatically overwriting the definitions in the CDR which differ from the wording found in the EVS would wipe out work intentionally performed in the CDR. For example:

Is this a desirable outcome?
A safer approach than a wholesale global change, blindly wiping out across the board any work intentionally performed in the CDR, would be to have a version of the report I have given you with checkboxes next to each Term/Concept pair, which would allow a terminology specialist to review the discrepancies and selectively choose the Term documents which should be updated using the information in the EVS. The script would then perform the updates and report back which changes were made successfully, and which failed (for example, because another user had the document checked out).
A companion report could be generated showing the drug concepts in
the EVS which have no corresponding CDR document (neither a match on a
CDR document with the same concept ID, nor a match of the normalized
preferred name with a CDR document which has no
NCIThesaurusConcept element, as reported above), with
checkboxes similar to the primary report for selecting concept which
should be imported as new Term documents in the CDR.
Finally, a third report could be created showing those CDR documents
with no NCIThesaurusConcept element, as reported above,
whose normalized preferred name matches the name of a concept in the EVS
which is not already associated with another CDR documents. Again,
checkboxes would be available to identify which of these CDR documents
should be updated with the information in the corresponding EVS
concept.
You might also want to have a fourth report comparable to the
spreadsheet I just attached showing concepts in the EVS which have been
attached to more than one Term document in the CDR. The
other reports obviously can't do anything with these documents until the
ambiguity is resolved.
~oseipokuw Do the other
name and definition statuses (Reviewed/Unreviewed) still have the same
significance they used to? I would think that if we use the form I've
described above, with the user approving updates for each term
explicitly, it would be reasonable to say that all of the other names
and definitions of the refreshed Term documents had been
reviewed, and could be marked as such. Do you agree?
You can preview the main form at https://cdr-dev.cancer.gov/cgi-bin/cdr/RefreshDrugTermsFromEVS.py. The
submit handler is a stub for now, but the form itself is pretty much
complete. The form takes a little under a minute to load, which is
pretty fast, when you think about how many concepts it has to load from
the EVS and how many Term documents have to be loaded and
parsed from the CDR. On RemoteApps Chrome appears to be truncating the
form partway into the "C"s, but Firefox and MS Edge have no problem with
it. ~volker reports that
Chrome works on his MacBook, so I assume CBIIT has crippled Chrome on
RemoteApps somehow, and it's not really a problem intrinsic to Chrome
itself.
~oseipokuw I noticed that of all 7,313 drug terms in the production CDR only one (CDR806181) had multiple definitions, and as far as I can tell the two definitions (including text, source, type, source ID, and status) are identical. Can I assume that this is a mistake? If I can assume that all correctly formed CDR drug terms have no more than one definition (as is true of all 7.482 concepts in EVS when I apply the rules for selecting definitions to be imported), then I may be able to enhance the form to highlight the differences between the definition we have and the definition the EVS has, which would surely make the reviewer's job much easier.
~oseipokuw I'd like to drill down on the requirements for normalization of names and definitions. Earlier you indicated that you wanted to ignore punctuation and capitalization, but made no mention of whitespace.
The approach I have taken so far has been to strip all characters
contained in the Python string.punctuation sequence, but
the drawback of using this approach is that it leaves some of the more
exotic "punctuation" character (with higher Unicode positions) in place.
I am in the process of refining this approach using some of the more
recent capabilities provided by the Python Unicode libraries for
distinguishing character classes, so I can truly preserve only
alphanumeric characters in the normalized versions of string for
determining "duplicates."
There are a number of approaches which can be taken to punctuation when normalizing the values.
The most conservative is to normalize whitespace (strip leading and trailing whitespace characters and collapse sequences of one or more whitespace characters into a single blank character) but to preserve punctuation in comparisons. With this approach, all three of the following names would be kept:
AZD 4017
AZD-4017
AZD4017
The first step away from that most conservative approach would be to remove all non-alphanumeric characters from the strings being compared, again with normalized whitespace. If the three names in the previous example were encountered in the order listed, then the following two names would be kept:
AZD 4017
AZD-4017
(The last name would be dropped because the normalized version of "AZD-4017" would be "AZD4017" which means that the software would concluded that we already have the third name.)
A variation of the previous approach would replace non-alphanumeric characters with a space and then perform the standard space normalization. With this approach the names to be kept would be
AZD 4017
AZD4017
The most aggressive normalization would strip all non-alphanumeric characters, including spaces. In our example only one version of the name would be kept:
AZD 4017
I assume that the CIAT staff is aware that changes in punctuation can alter the meaning of text, and that it has been decided that this risk is worth taking in exchange for the benefits of storing fewer names.
Please give some thought to which of the approaches described here should be used in this context.
A final note on the form: the lists of "other names" show all the variants present in the documents, some of which will be dropped as duplicative based on the normalization rules adopted. If you prefer, I can show only the variants in the right-most column which would actually be used in the CDR document, but I thought that in the interim seeing all the names would help clarify the issues as you consider the options and their implications for the normalization rules.
Can you remind me why names which are identical to the preferred name are sometimes also included as "other" names?
Also, it occurred to me to wonder what, if we are arbitrarily
discarding OtherName blocks because their normalized names
match those of another OtherName block which we've already
added to the document, would be the value of the type and source
information we store with the names which we happen to be keeping. Do we
still have any use for that information?
While waiting for answers to some of the questions above, I have implemented a version of the submit handler which takes the "OtherNames" at face value (not repeating the preferred name), and uses the third of the four approaches to normalization described above. The script is installed on the CIAT/OCC admin menu on CDR DEV.
Thanks! This is helpful!
Yes, I agree. That would be exactly how we would apply the statuses if the review was happening in the XMetal.
This looks great. It took a little longer than 1 min to load for me but I don't think it was too long. We will review and let you know if we have any questions.
I am working on a revised version of the report as requested, but I want to be sure you had noticed that the report you already have shows the preferred name whenever the CDR and the EVS don't match.
Yes, I noticed that but it will be good to provide the preferred names in all cases.
The reason it takes a bit longer to load the initial form than it did originally is that I am now caching the concepts fetched from the EVS, which reduces the time to re-draw the form when you have processed the queued updates you have selected to around 15 seconds. I did this because I want to reduce the temptation to click the BACK button, which would take you back to a stale form with misleading data.
Here's an overview of the questions we went through during this afternoon's status meeting.
What was the outcome of the discussion with Lori about whether we should use the "CTRP Terminology" concept as one of the starting points for pulling drug concepts from the EVS?
That discussion hasn't taken place yet. We decided that because the concepts pulled in by "CTRP Terminology" expands the retrieved set to several times the number of drug term documents we have in the CDR, using this "concept" to seed the retrieval set is probably inappropriate. William will decide whether to have the discussion with Lori or to just go with that provisional decision. He will add a comment recording what he did/decided.Does the explanation I gave above about why some documents are listed as "not found in the CDR" even though those documents can be found by looking up the drug name (they have the wrong concept code) make sense?
Yes.Do you agree that it is safer and more appropriate to use a form on which a reviewer can decide which CDR drug term documents should be refreshed from the EVS, rather than have a massive global change of thousands of CDR documents at once, wiping out any work which was performed to improve the imported drug terms?
Yes.Should we implement all four of the reports described in the comments posted above on November 22, beginning around 2pm?
Yes.Is the appearance of multiple identical definitions in a CDR
Termdocument a mistake?
We inadvertently skipped this question during the meeting, but I assume the answer is "Yes" because the duplicate definition was eliminated in the interim.Which of the four normalization algorithms described above (2021-11-24 11:36) should we use for names and definitions?
The most conservative algorithm (listed first), normalizing whitespace and eliminating case differences, but preserving punctuation. William will confirm this decision with Mary, now that he understands the explanations of the different approaches more thoroughly. He will post a comment to the ticket confirming or amending the decision, as appropriate.Should the form show all the names found in a concept document, or just the ones which we will keep in the refreshed CDR document?
This question will be largely moot if CIAT confirms their decision to use the most conservative normalization algorithm (though case differences will still present instances of this condition). If not, we will revisit this question. In any case, William has asked if we could highlight either the names which the CDR document already has, or the ones which the CDR document doesn't already have (he will post a comment letting us know which option CIAT would prefer).Why is the preferred name of a term sometimes duplicated as an "other" name?
Mary and William have been wondering the same thing. The tentative decision was that we will not continue this practice. William will post a comment confirming this decision.What is the value of the information accompanying the other names (e.g., name type and source information), when that information only belongs to one of a series of names which normalize to the same string and the choice of which name to keep is arbitrary?
Again, this question is less meaningful if we stick with the current decision to use the most conservative normalization algorithm, although differences in case can still present conditions in which a single name is randomly kept from a group which only differ in case. William, please confirm whether or not we should keep or drop this information.Is the current approach to highlighting the differences between the definition found in the CDR term document and the definition found in the EVS concept record satisfactory? (The form is only marking up deletions with strikethrough if there are no additions to highlight.)
Yes, the users agree that this approach is more readable than showing all deletions and additions.
I have tentatively implemented highlighting for other names in the
concept record which aren't in the CDR Term document as
other names, and other names in the CDR Term document which
aren't in the concept's set of other names. Not hard to reverse the
logic to highlight the names which do match between the CDR
document and the concept, if that's what you decide you prefer.
Also, the software has been modified to incorporate yesterday's decision about which normalization algorithm to use.
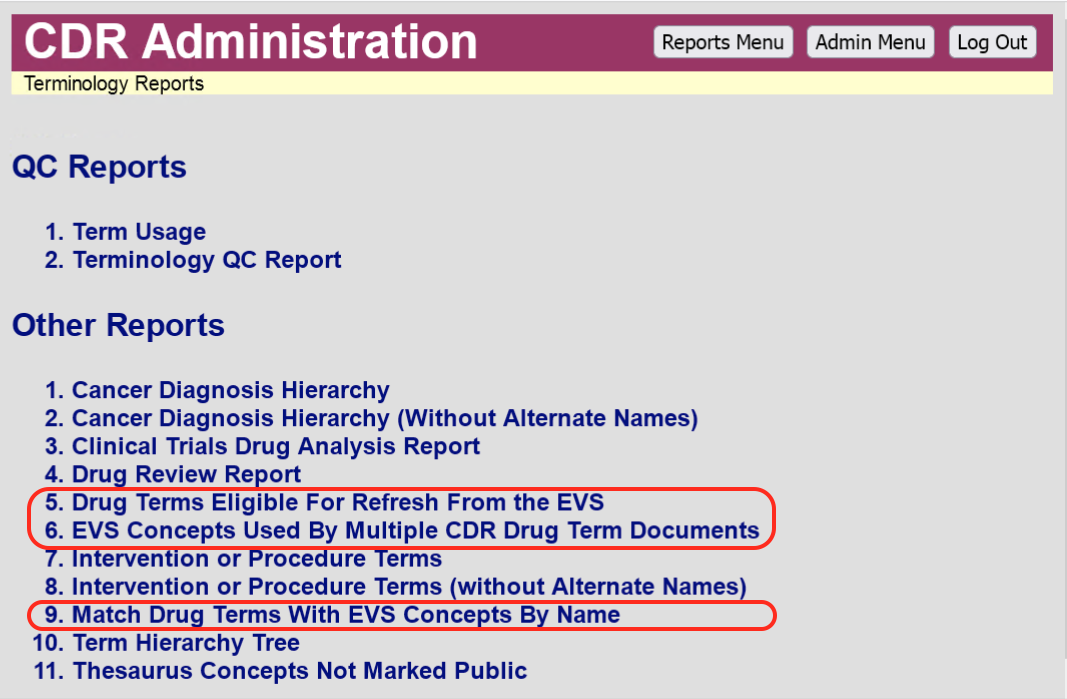
I have implemented the fourth report (EVS concepts used by more than on CDR drug term document).
https://cdr-dev.cancer.gov/cgi-bin/cdr/AmbiguousEVSDrugConcepts.py
I have moved this and the refresh form to the Term Reports menu, now that we have agreed that a form/report is preferable to a massive wholesale global change script.
Moving on to implementation of the other two reports.
For the other two reports, I need to find out which concepts haven't
been associated by concept code with any CDR drug term documents, but
match a CDR drug term document by name. As we have originally discussed
these reports, we have assumed that we would use the "name" property of
the concept records and the preferred name of the CDR drug term
documents (normalized for space and case) for this matching. The pitfall
for this approach can be illustrated by imagining the case of a drug
term which has two synonymous names, and the EVS picks one of those
synonyms as the preferred name, and the CDR picks the other synonym as
the preferred name. In that case, the interface which shows concepts for
which we can't find a match in the CDR either by code or by name, as
candidates for importing as new CDR drug Term documents
will include this drug, when what we'd really like to have
happen is that this drug shows up on the list of concepts which should
be used to refresh existing CDR documents which describe the
same drugs as their corresponding concepts. So this makes it tempting to
collect ALL the names for the concept records which aren't already
associated with CDR documents by code, and see if any of those names
match any of the names found in CDR documents without a concept code.
But here we run into another problem. It is true that the EVS ensures
that no two concept records have the same "name" property, even if we
ignore differences in case. (At least I assume that's true based on an
analysis of the 7,502 drug concepts in the EVS.) However, the EVS fudges
a bit when it comes to synonyms (and I haven't even started analyzing
the IND or NSC codes, or the CAS registry names for duplicates),
assigning the same "synonym" to two or more concepts. For example,
"GPOH-HD 95 Regimen" appears as a "synonym" for both C67163 ("OPPA
Regimen") and C67165 ("OEPA Regimen"). So even though Webster's defines
"synonym" as "a word that has the same meaning as another word in the
same language," this "synonym" represents two different concept records
with different definitions, and even a different drug combination for
the regimens described.
So here's what I propose.
For the form that shows EVS drug concepts not linked to any CDR term document by concept code, and for which we have found a match between on of the concept's names and one of the CDR term document's names, we will only include concepts NONE of whose names is found in another concept record, and we will only include CDR term documents NONE of whose names if found in another CDR term document.
For the form that shows drug concepts which are proposed as importable as new CDR term documents, we will only include concepts none of whose names appear in any other EVS drug concept record, AND none of whose names match ANY name found in ANY CDR drug document.
We add a fifth report, showing the drug concepts for which corresponding CDR documents have been identified, but which do not appear on the form showing concepts which can be imported as new CDR term documents, with the problem preventing that inclusion described (for example, "concept has name 'XXX' which is also found in concept C99999," or "concept has name 'XXX' which is found in CDR documents CDR123456 and CDR654321"). A second section of this report of anomalies will show all of the CDR drug term documents which have no concept code and which do not appear on the form showing CDR term documents matched by name to concepts which can be used to refresh those documents, with the reason why the documents were not included on that form (for example, "no names in this document match any names found in any EVS drug document," or "name 'XXX' in term document also found in CDR987654," or "name 'XXX' in term document matches concept records C12345 and C54321"). A human reviewer will need to sort out resolution of the cases in this report.
Please let me know when you and Mary have reviewed this proposed approach, ~oseipokuw, and whether you both agree with it.
As an aside, you haven't made any mention of such things for this
ticket, but you may want to consider simplifying the process and
eliminating some of the name/matching ambiguities by adding instructions
along the lines of "pretend that blocked CDR term documents don't exist"
or "ignore all CDR term documents whose term type is 'Obsolete term'" or
"ignore all OtherName elements having OtherNameType of
'Obsolete name'." Up to you.
Also, another incentive occurred to me for reviewing the proposed
term document refreshes instead of sending in the global change
bulldozer to wipe out any changes made by CIAT. In the existing
one-at-a-time interface for updating a CDR drug Term
document from a concept we have a safety measure in place which refuses
to perform the update if the preferred names don't already match. We're
not enforcing that in the new form, by which I mean we propose updates
for terms linked by code to concept records whose primary names don't
match the preferred names in the CDR Term documents. That's a good
thing, because it means we'll make the work of such updates easier, but
it gives the reviewer a chance to catch the cases where the wrong codes
were applied by mistake to the CDR documents. Hopefully, with all the
names and definitions displayed on the form such mismatches will be
obvious.
Because the logic needed for each of the remaining reports has so much overlapping commonality, I have combined them into a single page, added to the Term reports menu as Match Drug Terms With EVS Concepts By Name. This page has four sections.
A form, similar to the one used by Drug Terms Eligible For Refresh From the EVS, except that the CDR documents appearing in this form have no EVS concept code in the
NCIThesaurusConceptelement. Instead the pairs of documents appearing on this form are matched because they meet the following conditions: (a) no name in the concept record is found[*] in any other concept record; (b) no name in the concept record is found in any other CDR drug term document; (c) no name in the CDR document is found in any other CDR drug term document; (d) no name in the CDR document is found in any other concept record; and (e) at least one name found in the concept record matches one of the names found in the CDR drug term document. As with the original form, checkboxes are provided for the reviewer to select which pairs should be used to update the matching CDR documents with the values (including the concept code) of the corresponding EVS concept record.A form showing EVS drug concept records whose codes are not found in any CDR drug term document, and none of whose names are found in any other EVS drug concept record, nor in any CDR drug term document. Each concept record is accompanied by a checkbox for selecting the record for import as a new CDR drug term document. The concepts codes, names, and definitions are shown on the form.
A list of concepts which cannot be matched or imported, each with the explanation for the concept's problem preventing that match or import.
A list of CDR drug term documents which are not unambiguously matchable with any EVS concept, each with a description of the document's anomaly preventing such a match.
The processing logic for these scripts is fairly complicated, as you
can imagine. We've pretty much covered the universe of CDR drug term
documents and EVS drug concepts with these reports. Any EVS concept not
appearing on any of the reports has been associated (in the
NCIThesaurusConcept element) with exactly once CDR drug
term document, and the normalized names and definitions match. The same
is true for any CDR drug term document not appearing in any of the
reports. Because of this complexity, and the comprehensive nature of the
reports (and the opportunity for data corruption if any of these scripts
are flawed), it will be important that they are all thoroughly tested on
the lower tiers before promoting them to production. However, before you
dig too deeply into the testing efforts, I strongly suggest you provide
answers to the unresolved questions listed above, so I can first make
whatever adjustments to the logic are appropriate.
Shall we add this ticket to Ohm?
[*] When the description above says that a name is not found in concept records, we are ignoring names which are ignored when importing values into the CDR, such as names referring to hierarchical relationships to other narrower or broader concepts.
The new page take a little bit more time for the initial load than the one we reviewed on Thursday, because it's doing a little more work, but it's also using the same caching technique as that report, so follow-on pages for a session will take much less time.
~oseipokuw The case I was describing in this morning's PDQ meeting (CDR drug term document with lots more names than were contained in the matched concept record) no longer shows up on the form. I assume that happened as I tightened up the matching logic to conform to the rules described above for the form.
5. Is the appearance of multiple identical definitions in a CDRTermdocument a mistake?
We inadvertently skipped this question during the meeting, but I assume the answer is "Yes" because the duplicate definition was eliminated in the interim._
_
Yes, that was a mistake and it has since been corrected.
What was the outcome of the discussion with Lori about whether we should use the "CTRP Terminology" concept as one of the starting points for pulling drug concepts from the EVS?
That discussion hasn't taken place yet. We decided that because the concepts pulled in by "CTRP Terminology" expands the retrieved set to several times the number of drug term documents we have in the CDR, using this "concept" to seed the retrieval set is probably inappropriate. William will decide whether to have the discussion with Lori or to just go with that provisional decision. He will add a comment recording what he did/decided.
We decided not to discuss this with Lori since it is unlikely we will get any benefits on using the "CTRP Terminology" as one of the retrieval sources.
_
_
I have tentatively implemented highlighting for other names in the concept record which aren't in the CDR
Termdocument as other names, and other names in the CDRTermdocument which aren't in the concept's set of other names. Not hard to reverse the logic to highlight the names which do match between the CDR document and the concept, if that's what you decide you prefer._
_
The highlighting looks good. Thanks!
We would like to make a change to how the statuses are applied.
When a new term is imported, could you please add a new Review Status of "Imported" and if it is refreshed for existing terms, could you please add a new Review Status of "Refreshed" Also, in both cases, please do not make the document publishable as Mary would want to also review them in XMetal before making them publishable.
That will be a complete rewrite, as we would then no longer be able to use the global change libraries.
OK. Perhaps a report of terms that have been imported or updated should do, and we probably have one already for new or updated terms. We may have to get one or two of the existing reports modified if necessary. I will review the current reports and let you know if any of them needs updating.
Once again, I think we've been putting the cart before the horse. The fact that your original request was for a global change which would have updated thousands of documents at once, creating publishable versions with the changes for all of the terms which already have at least one existing publishable version, and now that request has morphed into asking for a system where you can not only review the refresh candidates and carefully select the ones to be processed, but also defer the creation of publishable versions with the changes so a second review can take place, leads me to think that I might not be the only one who's a little fuzzy on what the business goals are for the drug term documents. So let's first take a step back and articulate those so we can all be clear about what the relationship should be between the EVS and the CDR in this arena. Should one be essentially the authority and the other mirroring the values in the first? Or are they sharing the rôles? What are all the uses we make (and plan to make) with the drug term documents? What's the ideal state for the two repositories? Are there any unnecessary duplications of effort? Should there be more coordination between the EVS and the CDR? Less? Are we carrying information we don't need to maintain? Are we missing some information we need? And probably some other questions I've haven't thought of.
Yes, according to Mary don't show Obsolete and blocked terms.
In the meeting with Mary yesterday, we agreed to continue to use the global change harness for importing and updating the terms from EVS. Also, the two new requirements we talked about were:
1. For existing terms, Other Names with a Review Status of "Reviewed"
should not be overwritten when a document is refreshed. Also, Other
Names with a Review Status of "Reviewed" should not be dropped or
removed even if they do not appear on the corresponding concept from the
EVS.
2. When new terms are imported into the CDR, they should not be made
publishable.
Has Mary reviewed this language? Has she decided that it should read "... should not be overwritten ..." and not "... should not be overwritten or removed ..."? (In other words, if a name does not appear in the corresponding EVS concept, we will drop it from the CDR document.)
Yes, I assume you're saying that if the CDR document has an Other Name with a status other than "Reviewed" and that Other Name does not appear in the EVS concept, then it will be dropped. If that is the case, then yes, it is OK to drop it. However, if you're referring to Other Names that have a status of "Reviewed", even if they do not appear in the EVS concept, then they should be retained.
I have added ~barnsteadm as a watcher to the ticket.
If that's what you want, I suggest you amend the wording of the first of the two new requirements in your comment above accordingly.
Just a reminder note that ~oseipokuw or ~barnsteadm was going to try and find out what the cycle was for when EVS refreshed their exposed database. The plan is that we'll use that cycle to optimize the caching of concept records. If the cycle is unpredictable, or EVS won't disclose that information, our fallback plan was to implement an interface for refreshing our cache on demand.
Also, I have written confirmation above that Mary does not want obsolete terms, but I believe there was a verbal discussion which went further than that, instructing us to ignore all terms which do not have the term type of "Index term" (so, for example, we would ignore terms which have a term type of "Drug Combination" and do not also have a term type of "Index term"). Did I just imagine that? If not, please add a comment confirming that change.
On a separate note, based on ~oseipokuw's answer to my earlier question about blocked terms and obsolete terms and/or names, although I will be ignoring obsolete terms, I will not be ignoring obsolete names.
Also, I believe ~barnsteadm mentioned in last Wednesday's meeting on this subject that she wants us to keep the names she has marked as "approved." However, "Approved" is not one of the valid review statuses for other term names. The valid values are
Unreviewed
Reviewed
Problematic
This is correct. "Reviewed" generally means the term has been reviewed and "approved".
This is right. You did not imagine that 😃
Yes, this is OK.
Mary tried to find out about the EVS cycle but did not get a reliable response. It seems we cannot rely on their update cycle as it is not consistent.
I recommend that we drop the term import from the advanced term search so we don't have to maintain the logic in two separate placed.
~oseipokuw (or ~barnsteadm): Please create a single statement of the requirements for this ticket so that we will be able to determine whether the resulting implementation meets those requirements without digging through 67 different comments.
That would work for me.
Thanks!
Mary
From: NCI JIRA Mail (NIH/NCI) <NCIJIRAMBX@mail.nih.gov>
Sent: Tuesday, January 11, 2022 9:48 AM
To: Barnstead, Mary (NIH/NCI) [C] <mary.barnstead@nih.gov>
Subject: [NCI JIRA Tracker] Updates for OCECDR-5016: Importing multiple
terms from the NCI Thesaurus
There are 2 comments.
[1]OCPL Central Data Repository Support (CDR)
/
[2]OCECDR-5016
In Progress
Importing multiple terms from the NCI Thesaurus
[3]View issue · [4]Add comment
2 comments
Kline, Bob (NIH/NCI) [C] on Tuesday, 11 Jan 2022 09:36 AM
I recommend that we drop the term import from the advanced term search so we don't have to maintain the logic in two separate placed.
Kline, Bob (NIH/NCI) [C] on Tuesday, 11 Jan 2022 09:42 AM
[5]Osei-Poku, William (NIH/NCI) [C]: Please create a single statement of the requirements for this ticket so that we will be able to determine whether the resulting implementation meets those requirements without digging through 67 different comments.
This message was sent by Atlassian Jira (v8.13.4#813004-sha1:e00b0ab)
Jira is improving email notifications, [6]share your feedback!
Get Jira notifications on your phone! Download the Jira Server app for
[7]Android or [8]iOS.
----------------------------------------------------------------------------------------
[1] https://tracker.nci.nih.gov/browse/OCECDR
[2] https://tracker.nci.nih.gov/browse/OCECDR-5016
[3] https://tracker.nci.nih.gov/browse/OCECDR-5016
[4] https://tracker.nci.nih.gov/browse/OCECDR-5016#add-comment
[5] https://tracker.nci.nih.gov/secure/ViewProfile.jspa?name=oseipokuw
[6] https://surveys.atlassian.com/jfe/form/SV_aWUQ0lsYz9m8obb
[7] https://play.google.com/store/apps/details?id=com.atlassian.jira.server
[8] https://apps.apple.com/us/app/id1405353949
So are we in agreement that the software will use a cache which will be refreshed when explicitly requested using a separate interface? Or would you prefer that we stick with the current behavior (the user has to wait for a cache to be built when the page is requested from the menu, and that cache is used for the follow-on cycle of select-terms/process-terms/form-is-redrawn)? Or would you prefer that no cache is used and we fetch the records from the EVS every time the page is drawn?
Hi Bob-
i'm okay with waiting for the cache to be built.
Thanks,
Mary
FYI - I accidentally moved this ticket into the CGOV project while trying to move a different one. Amy was able to move it back into the CDR project as I did not have the permission to move it back. Sorry for the multiple notifications.
I have compiled the scattered requirements throughout this ticket below. Please make feel free to make any needed changes.
Summary of requirements for this ticket:
To identify all drug terms from EVS that are cancer related which we do not already have in the CDR, download them for review and potentially import them into the CDR.
If we already have the terms in the CDR, then check to see if there are any updates which we will review and import as well.
We decided to take the conservative approach to updates which meant capitalization would be ignored but not punctuation.
Add preferred names to the report, highlighting them so they stand out from the rest of the data.
Continue to use the global change harness for the reports. For existing terms, Other Names with a Review Status of “Reviewed” should not be overwritten when a document is refreshed.
Also, Other Names with a Review Status of “Reviewed” should not be dropped or removed even if they do not appear on the corresponding concept from EVS.
When new terms are imported into the CDR, they should not be made publishable.
Exclude obsolete terms (but not obsolete Other Names) from the report and ignore terms that are not index terms.
we only want to see true differences in preferred names and definitions not just capitalization or punctuation ...
I had requested a consolidation of the scattered requirements, not new changes to those requirements. I believe the most recent decision (captured in earlier comments above) as that we would ignore capitalization but not punctuation.
No, I would like to keep the punctuation as our style requires. Thanks!
Have you read this comment (in particular, #6), which summarized the decisions taken at the December 2 CDR/EBMS weekly status meeting? Did William not consult with you about it as he said he would?
6. Which of the four normalization algorithms described above (2021-11-24 11:36) should we use for names and definitions?
The most conservative algorithm (listed first), normalizing whitespace and eliminating case differences, but preserving punctuation. William will confirm this decision with Mary, now that he understands the explanations of the different approaches more thoroughly. He will post a comment to the ticket confirming or amending the decision, as appropriate.
The italicized portion refers to the decision captured from the meeting. The non-italicized part preceding it refers to the question where I was asking William to identify which of four possible approaches to normalizing strings for names and definitions we should take. The approach he identified in the meeting was the first one:
The most conservative is to normalize whitespace (strip leading and trailing whitespace characters and collapse sequences of one or more whitespace characters into a single blank character) but leave to preserve punctuation in comparisons. With this approach, all three of the following names would be kept:
AZD 4017
AZD-4017
AZD4017
Apologies for the multiple edits of the previous comment. Jira kept stripping the link markup from "Have you read this decision ..." so I had to come up with another way to link to the comment above.
Looks like Jira's internal linking is still broken. You'll just have to expand the collapsed comments and then scroll to the one timestamped "Thursday, 2 Dec 2021 03:59 PM" and then look at #6 in that comment.
Perhaps I read it incorrectly, but I would like to ignore differences in both capitalization AND punctuation between the two datasets, retaining the style that the PDQ has set.
While you're going back to read the original comments, let me try to pre-emptively clear up confusion about the language in those comments (definitely an on-target thing to do when we're talking about how to handle terminology documents).
The purpose of "normalizing" strings to be compared is to control how
we decide that string A is "different" from string B. In this context
"conservative" means "making as few changes to the strings being
normalized as possible." So at one end of the spectrum, the absolute
most conservative approach is to make no changes at all to the strings.
In this world, "French fries" and "french fries" are different, as are
"french fries" and "french
fries" (with a line break instead of a blank space between the two
words). The options I laid out back in November didn't even consider
that extreme an approach to preserving differences between strings.
At the other end of the spectrum, a really aggressive approach might uppercase all letters in the normalized string and then eliminate all characters except the ASCII letters A-Z. So "A75-b169:c(14')" would not be regarded as different from "abc" for purposes of comparing those two values. Or if we really wanted to get silly, we could same something like all strings having 20 or fewer characters get converted to "A" and all other strings become "B". Now that's about as far away from "conservative" as you can make normalization (without converting all strings to "A").
The further our algorithm moves to the conservative end of this
spectrum, the more frequently we will decide that our CDR
Term document needs to be updated, and the more names those
documents will retain on average. Moving to the other end of the
spectrum, using more aggressive normalization algorithms, we would end
up with fewer determinations that a CDR Term document is
different enough to need updating with values from the EVS, and fewer
aliases would be preserved. It would also mean that we increase the
number of false positives in the decisions the software makes when it
tries to match up CDR Term documents without a concept code
with EVS concepts which share one or more "normalized" names.
I hope this helps clarify what is meant when we describe a normalization as more or less "conservative" (the mnemonic here would be that the more "conservative" algorithms "conserve" more of the original string). I can understand how someone might get confused and think that "conservative" should mean "I get to keep as many of the capitalization and punctuation decisions I have made in the CDR as possible." But you'll want to remember that we've addressed that issue be a separate requirement which says "if a name has been marked in the CDR as Reviewed we keep it.
We should probably not do anything else with this ticket until we have achieved the state in which all of us understand the same thing when we read the requirements. If anything I have tried to explain here doesn't make sense to you, now is the time to say so. 🙂
Well, we've already established the requirement that we will keep every name which you have marked as Reviewed in the CDR. This means that if the only difference between what we have and what the EVS has is a name in the CDR which the EVS doesn't have but which you have asked us to keep, then no update will take place. What you're saying now is different from that requirement, as the retention of a name variant in the CDR would be required regardless of the name's review status.
And it's also different from the requirements established back in December, when William said we were to preserve punctuation when normalizing.
So in this latest version of the requirements, would "AZD4017" and "AZD-4017" be normalized to the same value? How about "AZD 4017" and "AZD-4017"?
I think the confusion stems from the fact that I included the statement from the original requirements (about capitalization and punctuation) instead of updating it with the later decision referencing the conservative approach to how the updates will be compared and implemented. I have updated the summary of requirements to capture that information. So, hopefully we are all on the same page.
Are we, ~barnsteadm?
OK, let's try it this way. I have attached a small spreadsheet (ocecdr-5093.xlsx) with cases which assume there are no differences in the definitions for given EVS concept/CDR drug Term pairs (though I'm assuming the same normalization algorithm — whatever that turns out to be — will be applied to definitions and to names). I have filled out the first couple of rows myself to get things started. The first two columns show the other names for the EVS concept record and the CDR Term document, respectively. Please fill in the third column for the remaining rows and reattach.
Sorry, I think I missed that in your last comment you switched to agreeing with the current version of the consolidated requirements as far as preserving punctuation during the normalization process, ~barnsteadm. So I think the only loose end to tie up is what we will do when names differ in case between the EVS and the CDR and the name in the CDR is not marked as approved ("Reviewed"). We have established that when the name is marked Reviewed we retain the CDR's version. Are you asking that we retain the upper/lowercase version found in the CDR for names which are not marked Reviewed?
If a name is not marked "reviewed", it can be removed or overwritten with the EVS name.
Thanks, I think I've got everything I need now to wrap things up.
OK, I think we're ready to declare dev complete for this one.
Verified on DEV. Thanks!
Verified on QA. Thanks!
| File Name | Posted | User |
|---|---|---|
| image002.png | 2022-01-11 10:00:06 | Barnstead, Mary (NIH/NCI) [C] [X] |
| image-2021-11-22-10-59-15-946.png | 2021-11-22 10:59:16 | Kline, Bob (NIH/NCI) [C] |
| image-2021-11-22-12-43-28-665.png | 2021-11-22 12:43:29 | Kline, Bob (NIH/NCI) [C] |
| ocecdr-5016.json | 2021-08-19 11:33:57 | Kline, Bob (NIH/NCI) [C] |
| ocecdr-5016.xlsx | 2021-09-16 12:27:05 | Kline, Bob (NIH/NCI) [C] |
| ocecdr-5016-ambiguous-codes.xlsx | 2021-11-22 16:21:38 | Kline, Bob (NIH/NCI) [C] |
| ocecdr-5016-codeless.xlsx | 2021-11-22 12:20:49 | Kline, Bob (NIH/NCI) [C] |
| ocecdr-5093.xlsx | 2022-01-20 07:16:29 | Kline, Bob (NIH/NCI) [C] |
| RE Automating transfer of new and updated drug terms.msg | 2021-09-27 17:28:51 | Osei-Poku, William (NIH/NCI) [C] |
| Screen Shot 2022-01-21 at 4.08.18 PM.png | 2022-01-21 16:18:44 | Kline, Bob (NIH/NCI) [C] |
Elapsed: 0:00:00.000578