CDR Tickets
| Issue Number | 5259 |
|---|---|
| Summary | [XMetaL] Error Message when loading Document |
| Created | 2023-06-29 15:38:45 |
| Issue Type | Improvement |
| Submitted By | Englisch, Volker (NIH/NCI) [C] |
| Assigned To | Englisch, Volker (NIH/NCI) [C] |
| Status | Closed |
| Resolved | 2024-12-19 14:24:48 |
| Resolution | Fixed |
| Path | /home/bkline/backups/jira/ocecdr/issue.349987 |
After starting XMetaL and after loading a CDR document I received a pop-up window with an error message:
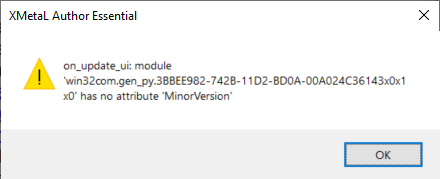
"on_application_open_complete: module 'win32com.get_py.3BBEE982-742B-11D2-BD0A-00A024C36143x 0x1x0' has no attribute 'MinorVersion'"
The following error message were displayed:
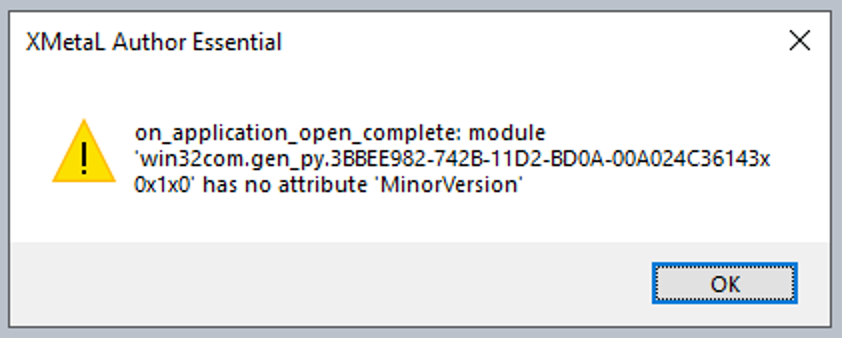
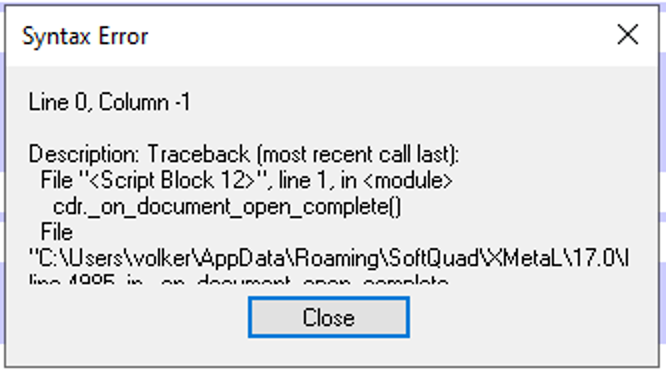
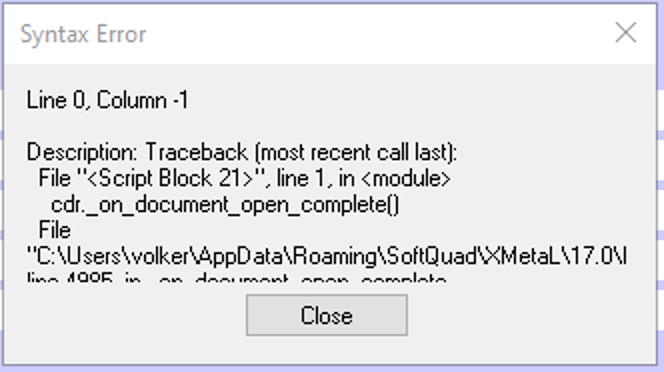
Adding ~oseipokuw as a watcher.
As mentioned in our status meeting, I saw these messages on a Monday morning after my machine was rebooted over the weekend with XMetaL open and running.
We were able to clean up the directory by removing some files/directories and restart XMetaL. Bob has written down the specific steps required for the clean-up.
From my notes taken during that session, here's a script which will generate the .CMD file which will fix a broken Python scripting registration.
import pathlib
import win32com
import win32comext
script = pathlib.Path(__file__).parent / "fix-python-scripting.cmd"
gen_py = pathlib.Path(win32com.__gen_path__).parent
client_dir =pathlib.Path(win32comext.__path__[0]) / "axscript" / "client"
with open(script, "w", encoding="utf-8") as fp:
fp.write(f'RMDIR /S /Q "{gen_py}"\n')
fp.write(f'CD /D "{client_dir}"\n')
fp.write("python pyscript.py\n")
fp.write("PAUSE\n")
And here's a wrapper script to let the user avoid opening up a
command console, running the python script, and then running the
generated .CMD file. This gets dropped into the CdrSetup17
folder (as {}fix-scripting-wrapper.cmd{}) alongside the
Python script above (named {}make-scripting-fixer.py{}).
The user right-clicks on the .CMD file and runs it with elevated
privileges, as with the other setup scripts.
REM @ECHO OFF
CD /D "%~dp0"
python make-scripting-fixer.py
CALL fix-python-scripting.cmd
The reasons we cant just do all of this in a single Python script are:
users are more used to launching programs from Windows Explorer than running Python scripts in a command console window
replacing broken registration from a running Python script would likely be blocked on Windows, which would complain that we're trying to replace things which are in use (works in Unix, but not in Windows), so we let Python figure out what needs to be done and write those instructions into a separate script to be launched after the Python script finished
Power broker supports running .CMD files with elevated permissions, but won't allow the same thing for running a Python script directly (of course, that's a little silly, since the .CMD file is allowed to run Python scripts, but that's how it works)
Who knows, we may never need this, but best to be prepared in advance. 😃
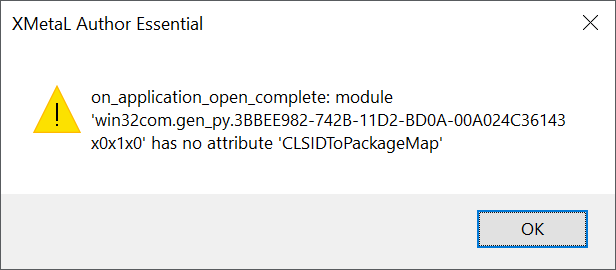
Leisha ran into a similar (but not identical) error message. Hers said:
on_commandbars_activate: module 'win32com.gen_py....' has no attribute 'CLSIDToPackageMap'
In her case, the problem resolved itself and she was able to work again after a reboot connected with a CBIIT BigFix update.
Leshia reported another problem this afternoon but once again after running a pending Big Fix installation and restarting her laptop, the message did not come up again.
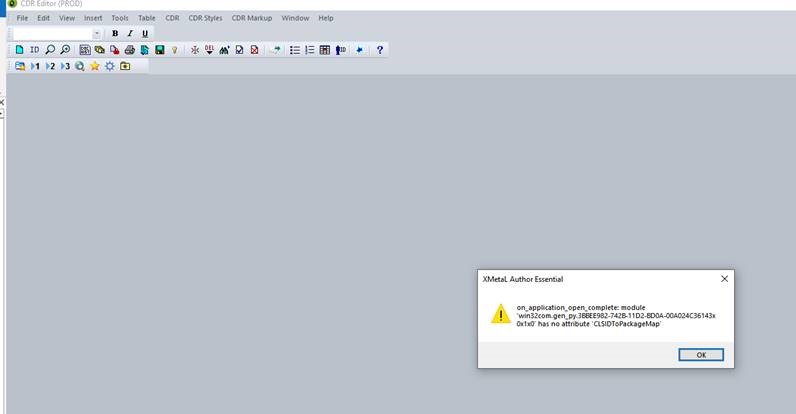
This may not be related to the other error message but I'll add a screenshot here to keep a record:
I had loaded a couple summary templates on DEV and closed those (without changes) followed by closing XMetaL when I saw this message.
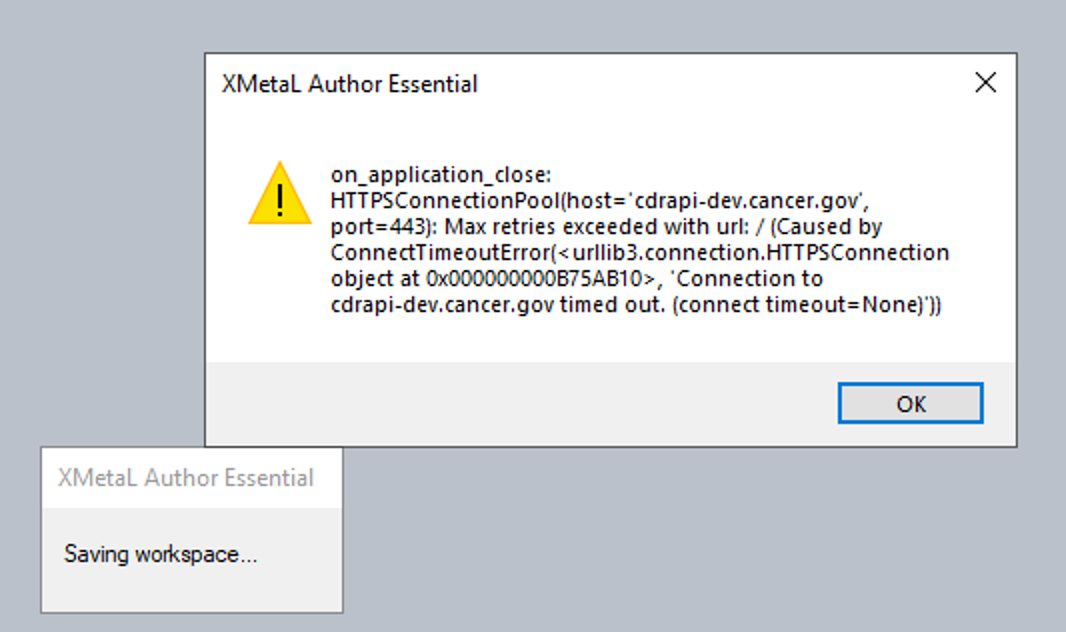
After clicking "OK" the warning message disappeared and XMetaL closed.
If I had to guess I'd say this is likely completely unrelated. The original problem is caused by corruption (or something like corruption) in the registration of the scripting language. In this case the scripting engine seems to be intact, and we're instead dealing with a failed TCP/IP connection. It's possible that the fact that the server you were trying to talk to was grinding away on an hour-and-a-half global change job made it harder for that server to keep up with other incoming requests. ¯_(ツ)_/¯
I wasn't aware you were bringing the server down to its knees with a global. That was likely the problem although I had never experienced such a message.
Well, this is the CBIIT network, so doesn't take much. 😃
I experienced a similar problem as Volker this afternoon on PROD at 1:49PM. I was trying to retrieve a document but instead of the document opening, I got this error message and XMetal froze. I had to close XMetal, start it again before I was able to do anything.
There have been quite a few reports of problems with retrieving network resources when on the VPN in past several days. In any case, I don't think we want to clutter this issue (which has to do with local configuration problems) with network failures. Please create a separate ticket to track the network connection failures.
Leshia reported a similar problem on 11/01/2023 - Restarting her computer resolved the problem.
Robin experienced a similar problem on 1/2/2024. Restarting her computer resolved the issue.
Judy ran into a similar problem this morning when he checked out a summary and tried to edit it. Once again restarting her computer resolved the issue.
We decided to close this ticket in last week's CDR Meeting.
| File Name | Posted | User |
|---|---|---|
| https connection error 1 49.PNG | 2023-09-12 18:21:18 | Osei-Poku, William (NIH/NCI) [C] |
| judy screenshot.png | 2024-02-16 11:24:47 | Osei-Poku, William (NIH/NCI) [C] |
| Leshia_screensht.PNG | 2024-02-15 12:53:50 | Osei-Poku, William (NIH/NCI) [C] |
| Leshie_on_application_cdr_error.jpg | 2023-08-30 16:47:46 | Osei-Poku, William (NIH/NCI) [C] |
| robin_screenshot.png | 2024-02-15 12:56:20 | Osei-Poku, William (NIH/NCI) [C] |
| Screenshot 2023-06-26 at 11.39.02 AM.png | 2023-06-29 15:40:58 | Englisch, Volker (NIH/NCI) [C] |
| Screenshot 2023-06-26 at 11.40.07 AM.png | 2023-06-29 15:41:19 | Englisch, Volker (NIH/NCI) [C] |
| Screenshot 2023-06-26 at 11.44.24 AM.png | 2023-06-29 15:41:36 | Englisch, Volker (NIH/NCI) [C] |
| Screenshot 2023-09-11 at 3.49.41 PM.png | 2023-09-11 15:53:21 | Englisch, Volker (NIH/NCI) [C] |
Elapsed: 0:00:00.001221